Introduction to Data on Imprinted Genes
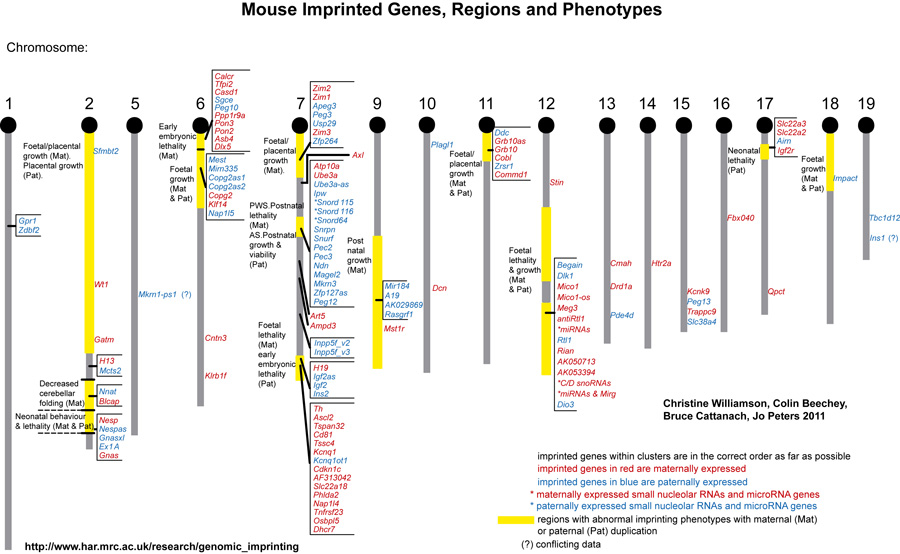
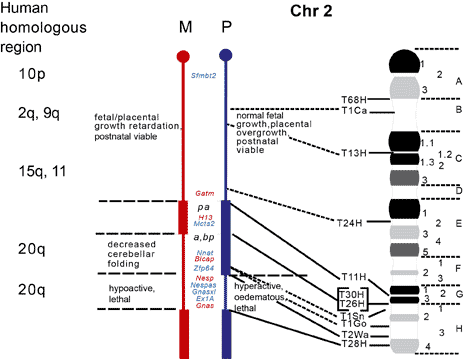
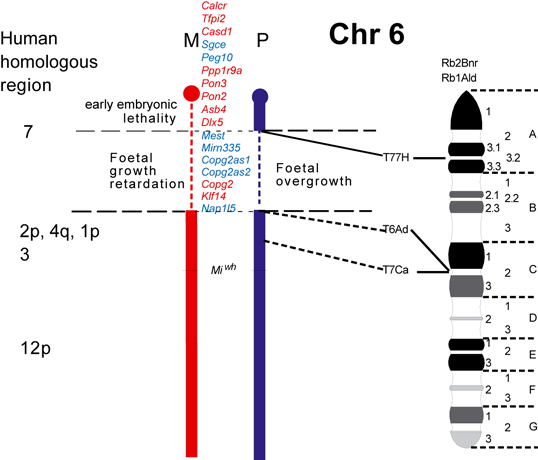
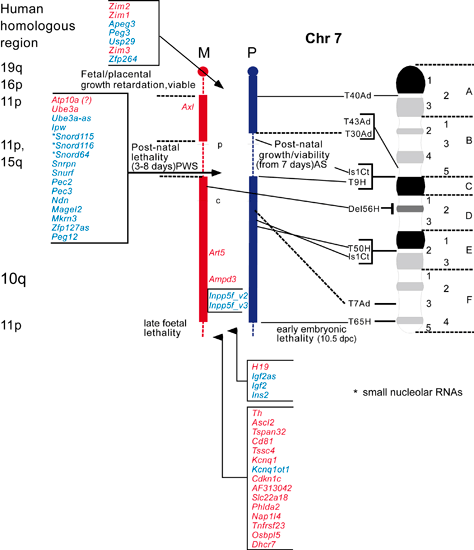
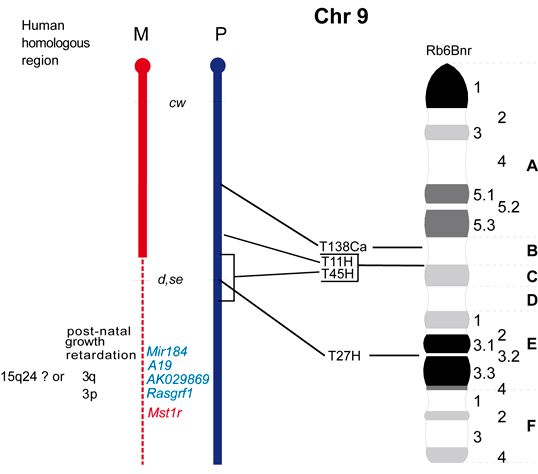
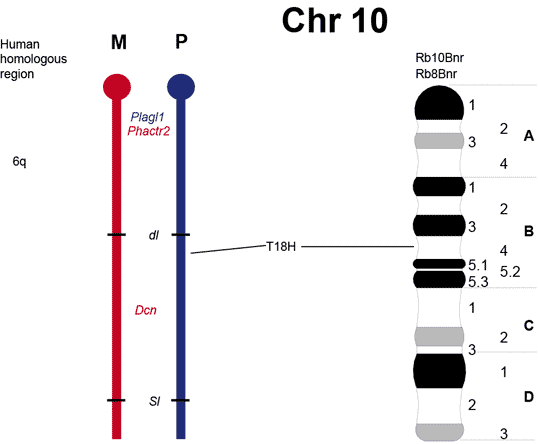
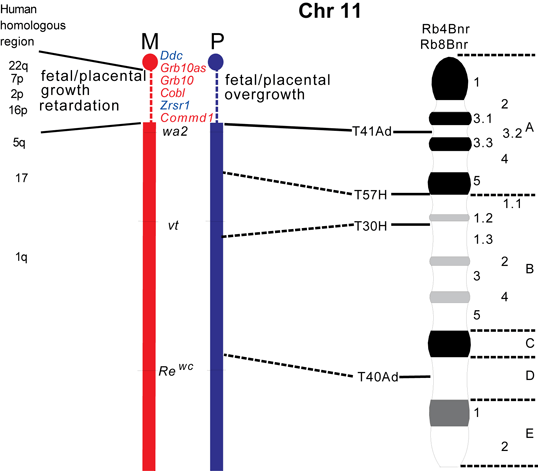
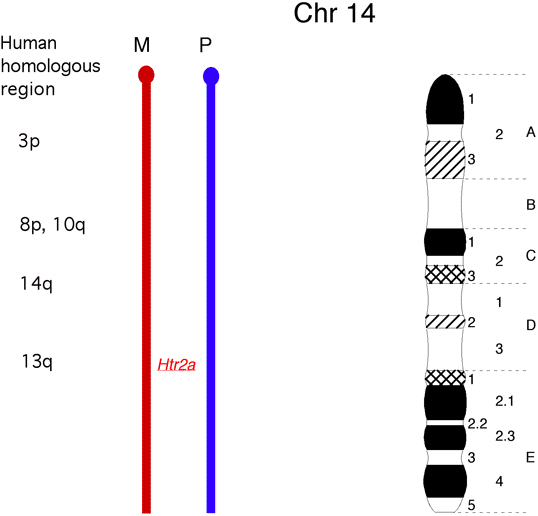
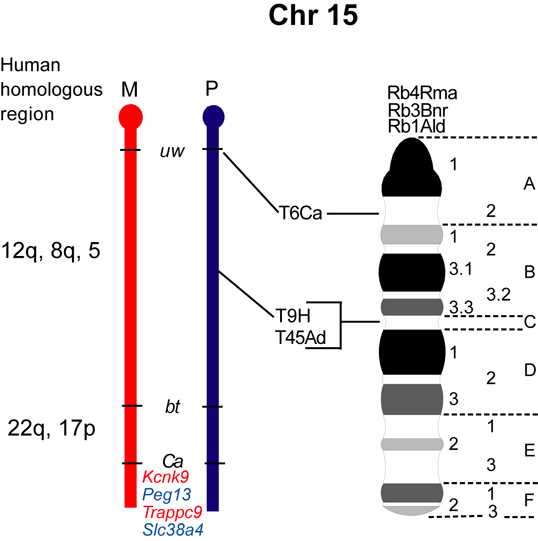
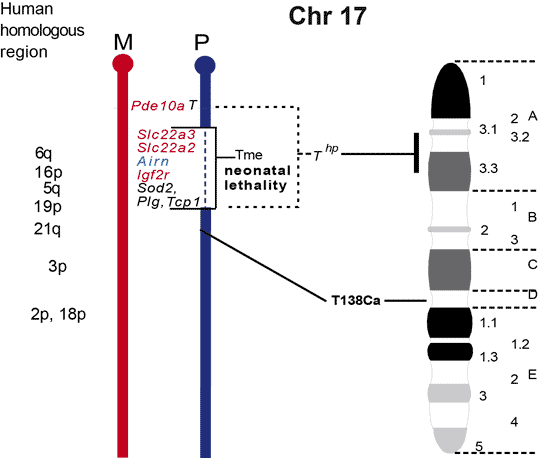
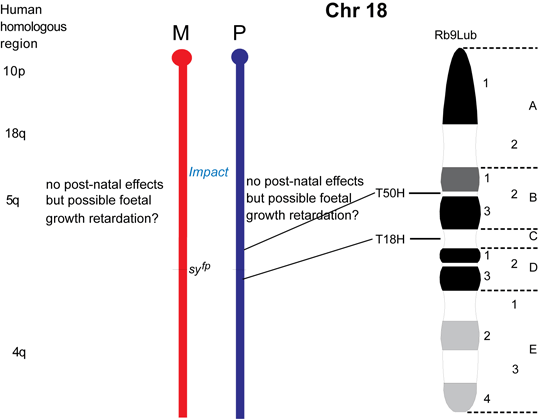
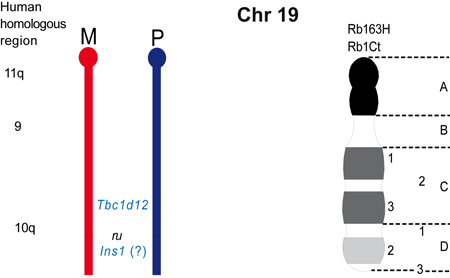
The maps were formerly published in Mouse Genome and are based on mouse genetic studies in which Robertsonian (Rb) and reciprocal (T) translocations have been used to generate uniparental disomies and uniparental duplications of whole or selected chromosome regions, respectively. The approved Nomenclature Rules applicable to these genetic studies are also given. Chromosome regions exhibiting imprinting effects (phenotypes) with 2 maternal (M) and no paternal, or 2 paternal (P) and no maternal copies are illustrated on both the ideogram, Evans (1996), Beechey & Evans (2004) and genetic maps. These maps are not exhaustive as some imprinting effects may be subtle. Imprinted genes within domains are not necessarily shown in the correct order relative to each other or to chromosomal orientation (centromere to telomere).
Other areas of the genome where imprinted genes have been identified are also shown, (chromosome 1, 5, 10, 13, 14, 15 and 19), despite no obvious abnormal phenotype being associated with maternal or paternal duplication.
The ideograms present the known imprinting status, up to 2011. This includes imprinted genes, imprinted small nucleolar RNAs (snoRNAs), microRNAs, (miRNAs) and their expressed parental allele. These data have been identified on the individual maps, along with the chromosome anomalies used to define each imprinting region. Gene symbols written in red are expressed from the maternal allele and those in blue from the paternal allele. MGI gene symbols are used. Each imprinted gene has links to the appropriate reference(s). Symbols for Robertsonian (Rb) translocations used to identify imprinting phenotypes are shown above each chromosome. The positions of reciprocal translocations (T) which currently define the regions with imprinting phenotypes are shown in bold with their positions, if known, on both the linkage and physical maps. Other reciprocal translocations, insertions (Is) and deletions (Del) that have been used to define or investigate imprinting regions are also identified.
Evans EP (1996) Standard Idiogram. In Lyon MF, Rastan S & Brown SDM (eds.) Genetic Variants and Strains of the Laboratory Mouse, 3rd ed. Oxford University Press: pp 1446-1448
Beechey CV & Evans EP (2004), MRC Harwell, Harwell, Oxfordshire. Standard Ideogram/Anomaly Breakpoints of the Mouse (URL: http://www.har.mrc.ac.uk/).
Lyon MF, Cocking Y & Gao XQ (1997) Mouse Genome 94:29-77